



Next: redo_mlrgd and redo_plots
Up: usersguide
Previous: Error log file
Contents
Parameters
There are a variety of parameters that you can alter by opening the
script run_mlrgd in a text-editor and editing the Options
section. A brief description of these are given below (suggested
defaults are give by param = default).
- pairs = 1: Method for generating the list of sequence pairs to
use: 2
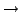 use phylogenetic tree built with ednapars (maximum
parsimony); 1
use phylogenetic tree built with ednapars (maximum
parsimony); 1 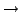 use phylogenetic tree built with ednaml
(maximum likelihood); 0 or other
use phylogenetic tree built with ednaml
(maximum likelihood); 0 or other 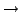 use user-input file
prefix.pairs.
use user-input file
prefix.pairs.
- refpos = 0: 0
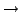 use seperate CDS files for each
sequence; 1 or other
use seperate CDS files for each
sequence; 1 or other  use reference sequence CDS files for all
sequences.
use reference sequence CDS files for all
sequences.
- gbkpos = 0: 0
 get CDS files from
seqname.fasta.orfs (or seqname.gbk.orfs) files; 1
or other
get CDS files from
seqname.fasta.orfs (or seqname.gbk.orfs) files; 1
or other  get CDS files from seqname.gbk files
(for any seqname.fasta files, the seqname.fasta.orfs
file, if any, will be used).
get CDS files from seqname.gbk files
(for any seqname.fasta files, the seqname.fasta.orfs
file, if any, will be used).
- skip = 0: 1 or other
 skip sequence files
containing non-ACGTUacgtu characters (exits if the reference sequence
contains non-ACGTUacgtu characters); 0
skip sequence files
containing non-ACGTUacgtu characters (exits if the reference sequence
contains non-ACGTUacgtu characters); 0  read standard
ambiguous nt codes as N and ignore for most statistics (ambiguous nt
are logged along with the gaps; codons containing ambiguous nt are logged
along with the `zero-probability' transitions).
read standard
ambiguous nt codes as N and ignore for most statistics (ambiguous nt
are logged along with the gaps; codons containing ambiguous nt are logged
along with the `zero-probability' transitions).
- window1 = 5:
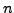 , where the running mean sliding window size
for the 1st, 2nd and 3rd codon positions and for the 4-fold degenerate
neutral sites plots is
, where the running mean sliding window size
for the 1st, 2nd and 3rd codon positions and for the 4-fold degenerate
neutral sites plots is 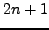 nt (or codons).
nt (or codons).
- window2 = 10:
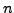 , where the running mean sliding window size
for the all sites and for the non-coding sites plots is
, where the running mean sliding window size
for the all sites and for the non-coding sites plots is 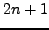 nt.
nt.
- circular = 1: 0
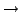 not a circular genome; 1 or
other
not a circular genome; 1 or
other 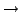 circular genome.
circular genome.
- clip1 = 0.00: Upper clipping threshold for clipped running
mean: clip1 = 0.05 means that the upper 5% (i.e. columns with low
observed number of mutations relative to expected number of mutations) in
each window are clipped before calculating the mean.
- clip2 = 0.30: Lower clipping threshold for clipped running
mean: clip2 = 0.05 means that the lower 5% (i.e. the columns with
high observed number of mutations relative to expected number of mutations)
in each window are clipped before calculating the mean.
- tttmin = 0.0: Lowest
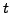 value to use in
value to use in 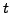 -fitting.
-fitting.
- tttmax = 10.0: Highest
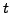 value to use in
value to use in 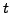 -fitting.
-fitting.
- tVfit = 0.2: Required fitting accuracy (total number of mutations
between a sequence pair) for
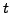 and
and 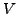 .
.
- maxiter = 20: Maximum number of fitting iterations for
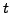 and
and 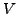 .
.
- Vmin = 0.01: Lowest
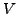 value to use in
value to use in 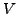 -fitting (
-fitting (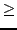 0.01).
0.01).
- Vmax = 10.0: Highest
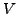 value to use in
value to use in 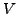 -fitting (
-fitting (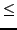 10).
10).
- Vtype = 1: 0
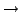 find seperate
find seperate 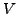 value for each
sequence pair; 1 or other
value for each
sequence pair; 1 or other 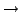 find one
find one 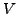 value for the alignment.
value for the alignment.
- Vfix = 0: If
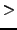 0, use this
0, use this 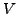 value for all sequence pairs
(defaults to 1 if
value for all sequence pairs
(defaults to 1 if 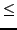 0; overridden if fitwhat = 3).
0; overridden if fitwhat = 3).
- fitwhat = 3: 0 or other
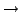 fit
fit 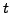 using total number
of mutations; 1
using total number
of mutations; 1 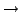 fit
fit 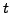 using number of neutral/synonymous
mutations; 2
using number of neutral/synonymous
mutations; 2 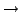 fit
fit 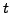 using number of neutral/synonymous
mutations at 4-fold degenerate sites; 3
using number of neutral/synonymous
mutations at 4-fold degenerate sites; 3 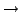 as for 2, then adjust
as for 2, then adjust
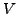 to fit the total number of mutations (i.e. including nonsynonymous
mutations).
Note: 1--3 are not recommended if the sequence is predominantly double-coding
since, in this case, the number of neutral sites is potentially very small.
to fit the total number of mutations (i.e. including nonsynonymous
mutations).
Note: 1--3 are not recommended if the sequence is predominantly double-coding
since, in this case, the number of neutral sites is potentially very small.
- wholeseq = 1: Nucleotide range to use for all model-fitting,
statistics and plots: 0
 use the region
range1--range2, 1 or other
use the region
range1--range2, 1 or other  use the whole sequence.
use the whole sequence.
- range1 = 0: Start nucleotide of region to analyse
(if wholeseq = 0; otherwise ignored), in reference sequence coordinates.
- range2 = 0: End nucleotide of region to analyse
(if wholeseq = 0; otherwise ignored), in reference sequence coordinates.




Next: redo_mlrgd and redo_plots
Up: usersguide
Previous: Error log file
Contents
aef
2007-12-10